Nanopore Detectors
What are Nanopore Detectors?
Nanopore detectors are instruments built around a tiny pore in a membrane or thin, solid-state wafer. The pore is just big enough to allow a single strand of DNA to pass through. A voltage applied across the membrane generates an ionic current capable of pulling DNA molecules (which are negatively charged) through the pore. When the DNA molecule blocks the opening of the nanopore, it causes a characteristic decrease in the current, allowing discrimination between individual DNA nucleotides. A computer trained by machine-learning techniques recognizes the signals generated by different DNA molecules.
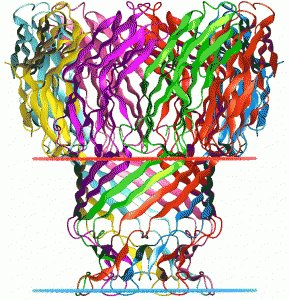
Protein nanopores can be constructed using a class of molecules, termed porins. We have previously used a nanoscale pore formed by the bacterial toxin, alpha-hemolysin(see below), and have recently been using a porin from Mycobacterium smegmatatis. The organism uses the porin for importing nutrients from its environment, but when the porin is placed in a lipid bilayer, it becomes well suited as a nanopore with dimensions that are particularly useful for interpreting the sequence of a single strand of DNA.
Controling the rate that DNA moves through the pore is essential to resolving individual DNA bases. Biological molecules that interact with DNA are well suited to this task. Examples include:
- RNA polymerase, which reads DNA in order to accurately make RNA transcripts;
- Exonucleases, enzymes that digests DNA starting at one end; or
- DNA polymerase, a class of enzymes that copy DNA.
We have evaluated DNA polymerases from Escherichia coli, and recently we have been working with a DNA polymerase from bacteriophage Φ-29.
Here is a depiction of the process showing single stranded DNA moving through the nanopore under the control of a DNA polymerase. As shown, bases that pass through the nanopore produce characteristic signal corresponding to individual bases along the strand.
Another porin molecule that we have used is from Staphylococcus aureus. This is the alpha hemolysin pore (3ANZ.pdb, Tanaka et al. 2011 Protein Science 20:448-456).
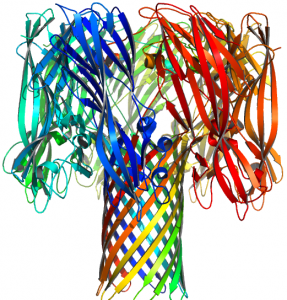
The base of the porin is inserted in a lipid bilayer in the cell, and when we use it as part of the nanopore device, it is placed in an artificial lipid bilayer. This single nanopore separates two chambers containing a buffered salt solution. The nanopore then becomes the only interconnecting path between those two chambers. Electric potential between the two chambers can be modulated, and the resulting current flow can be measured. When a DNA molecule enters the pore, the resistance increases, resulting in a decrease in ionic current through the pore. As individual nucleotides pass through the pore, characteristic changes in ionic current amplitude can be monitored.
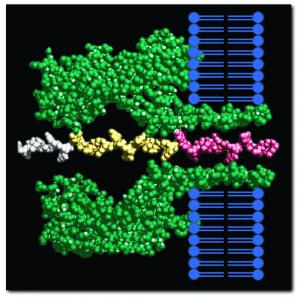
In side view, the porin complex penetrates the lipid bilayer (blue), allowing a single polymer chain of DNA to move between the two chambers separated by the lipid bilayer
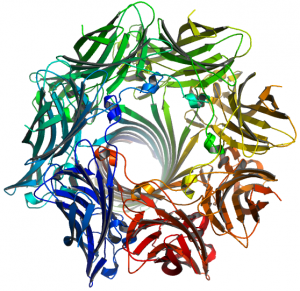
This top view of the alpha-hemolysin pore shows that it is constructed from seven different peptide chains.
